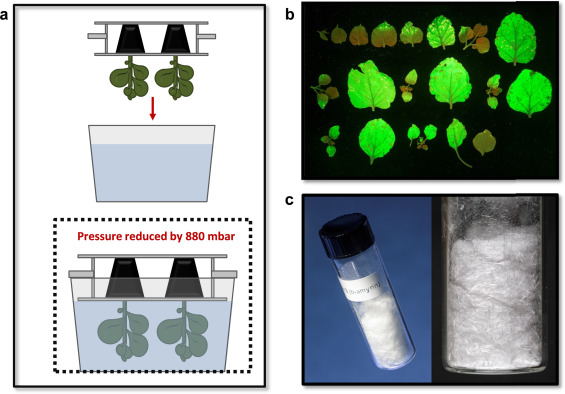
Guest blog from Emma McKechnie-Welsch, a PhD student from the John Innes Centre who spent three months doing an internship in Science Engagement with OpenPlant and the SAW Trust.
Plant Science SAW projects at Tunstead primary School
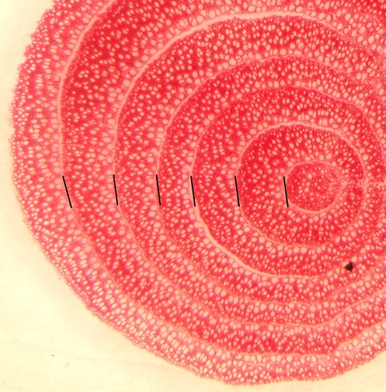
Arabidopsis apical meristem. Image by Emma McKechnie-Welsch
My name is Emma and I am a PhD student working in the Cell and Developmental Biology department at the John Innes Centre. My research looks at genes functioning to facilitate controlled plant growth and development from the shoot apical meristem in Professor Robert Sablowski’s research group. My PhD funding from the BBSRC includes a three month work placement and I was keen to gain experience in science communication and outreach so arranged a joint placement with OpenPlant and the SAW trust.
On my placement I had the opportunity to design two SAW projects to discuss science relevant to my research with primary school children at Tunstead primary school. For the year 1/2 class I worked with writer Julia Webb and artist Lara Nicole and the aim was to get children thinking about the functions of different parts of a plant. For the year 5/6 class I was worked with writer Mike O’Driscoll and artist Chris Hann with a day themed around plant evolution.
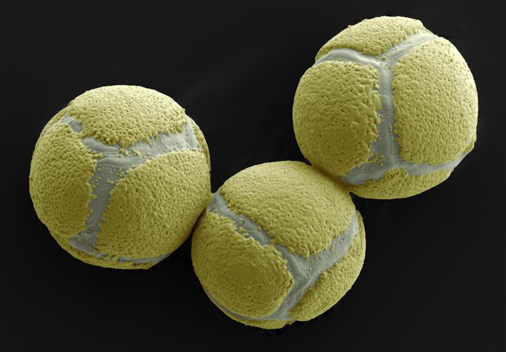
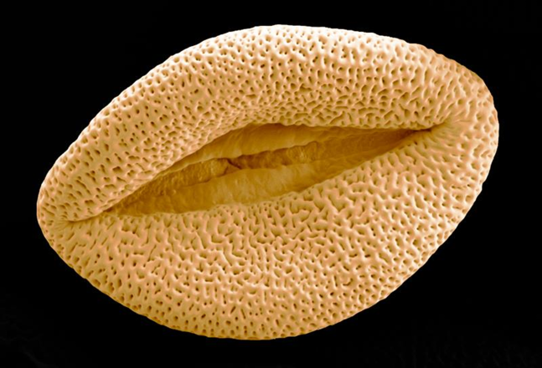
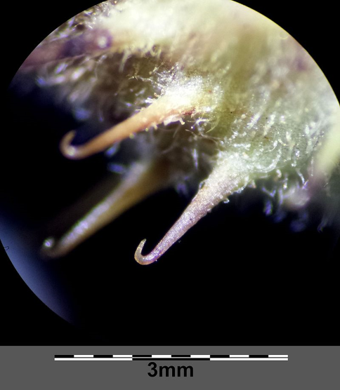
We used scientific images at the start of the day to catalyse inquisitiveness about the science we were going to explore, and provide inspiration for the poetry and art sessions.




Practical Science with Year 1/2
Build a plant game, played with year 1/2 class
To start off the lesson we played a “build a plant” game to get more familiar with the main parts of a plant, their function, and what plants use from their environment to grow. Each child also put a cut flower in coloured water to think about the use of the stem. Then the children were given a selection of fruit and vegetables and asked to decide what part of the plant each came from. They were given a flower to look more closely at the reproductive parts and think about how seeds are formed by pollination. Finally, they looked at different types of seeds in a seed kit and we discussed the different types of seed dispersal tactics plants use.
Practical Science with Year 5/6
The children dissected plants to look up close at the reproductive parts under the microscope.
We began by guessing the number of different plant species on earth and the children suggested why plants are useful. In groups, they were given cards representing each component of photosynthesis and had to arrange them to think about the process. We covered pollination and its importance for increasing genetic variation.
The children dissected plants to look up close at the reproductive parts under the microscope. I covered different types of seed dispersal and the importance of varying environmental conditions for evolution. Then children carried out DNA extraction from strawberries after learning a bit about what DNA was and how important it was in controlling the appearance of the plant, with a single mutation in a gene coding region potentially greatly changing this. Following on from DNA extraction there was a game to match the numbers of genes to different organisms.
After the morning science sessions the children had poetry and art sessions based on the content. Here are some poems and images from the Year 5/6 group (age 10/11):
The Year 1/2 children (age 4/5) wrote poems as if they were a seed growing up, and made flower hand puppets after designing a flower:
The children really engaged with the scientific learning aspect of the day which was great. Lots of the children thought about the questions I asked to the classes and gave insightful answers, as well as wanting to ask questions throughout the lesson/ activities. When asked about their favourite part of the day, at least half the children listed specific sections of the science morning.
The poems produced by the year 5/6 children really showcased the children’s interest in understanding genetics and how growth and development of organisms are controlled. The younger children were enthusiastic about looking at different types of seeds, bringing back different types they had found in their school grounds at break time to show me. It was great for them to think about the different stages of growth a plant goes through from seed to eventually producing a flower, including difficulties different environmental conditions could cause, while writing their poems.
The children were really excited about getting to do an afternoon of art although the activities designed weren’t quite as expected. The art didn’t centre around drawing on paper but producing 3D art pieces. The younger children gave lots of personality to their individual hand puppets and used them to help communicate their poetry whilst the older children focused on the scientific pictures provided and gave interpretations of pollen and seed dispersal, as well as the protective mechanism of the cactus.
From this experience, I could see how integration of science with writing and art can help children associate science more closely with creative thought, rather than a regimented, inflexible learning process, which makes the subject inaccessible to some children. The teachers were impressed with the pieces the children managed to produce and the level of thought about scientific processes they reached, which I think was largely down to the different approach to education SAW days take.

![[Closes 24 Nov 2107] Apply now to the OpenPlant Fund!](https://images.squarespace-cdn.com/content/v1/54a6bdb7e4b08424e69c93a1/1509564315902-TUO4I6QRWI9TT8UGSIAJ/OpenPlantTwitter_400x400+%281%29.jpg)

![[Closes 7 Mar 2017] OpenPlant Research Associate (Haseloff Lab)](https://images.squarespace-cdn.com/content/v1/54a6bdb7e4b08424e69c93a1/1486552818859-FH76MCA8SMFU93WB85RX/OpenPlantTwitter_400x400.jpg)














![[Closes 18 July 2017] OpenPlant PDRA: synthetic biology of riboswitches for plant and algal biotechnology](https://images.squarespace-cdn.com/content/v1/54a6bdb7e4b08424e69c93a1/1499679736557-UTX6PI913FZNBJ85W36D/OpenPlantTwitter_400x400.jpg)